Characterization of virulence gene distribution and antibiotic susceptibility profiles of diarrheagenic Escherichia coli from chicken faeces
Abstract
The inappropriate use of antibiotics in the poultry industry for prophylaxis and growth promotion has contributed to the development and spread of antibiotic resistance in zoonotic pathogens. This study aimed to understand the prevalence of pathogenic Escherichia coli (E. coli) in chicken faeces and their antibiotic susceptibility profiles. Forty-five E. coli isolated from the chicken faeces were further tested for virulence genes associated with diarrheagenic E. coli and extended-spectrum β-lactamase (ESBL) genes blaCTX, blaTEM, blaSHV and Metallo-beta-lactamase gene blaNDM. Results showed that eaeA and astA genes were detected in 60% of the isolates, followed by bfpA in 57.8%, lt in 26.7% and st gene in 2.2% of the isolates. Further, 15.5% of the isolates produced ESBL phenotypically whereas genotypically, one isolate was positive for blaCTX, one for blaTEM, and one for the blaSHV gene. Twelve isolates (26.7%) harbored gene blaNDM. High levels of resistance were observed against cefepime (93.3%), ertapenem (78.9%), meropenem (73.6%), tetracycline (88.9%), and cotrimoxazole (75.6%). This study highlights the importance of poultry as a source of human pathogenic multidrug-resistant (MDR) E. coli and their environmental dissemination.
INTRODUCTION
Escherichia coli is a common inhabitant of the intestinal tract of humans and animals and can be easily disseminated into different ecosystems through the food chain and water supplies [1, 2]. E. coli, being a part of the endogenous microbiota of animals and birds, can acquire resistance against antibiotics used in livestock [3]. The poultry industry is one of the fast-growing food production businesses worldwide. It is estimated that the global poultry population will reach 8.5 billion by 2030 [4]. Poultry meat is the low-cost common source of animal protein among the meat-consuming population. Hence, to fulfil their increasing demand, animal husbandry has met with extensive use of antibiotics [5]. The global consumption of antibiotics used for poultry will increase by 67%, from 63,151 tons in 2010 to 105,596 tons in 2030, primarily by BRICS (Brazil, Russia, India, China, and South Africa) countries [6]. The major reason behind this increase is the unregulated use of antibiotics employed to promote poultry growth and to control various infectious diseases in the poultry industry [7]. As a result, there is a high antibiotic selection pressure leading to bacterial resistance in poultry and consequently, their faecal flora contains a relatively high proportion of resistant bacteria [8-10].
The drug-resistant and animal intestinal bacteria such as E. coli can be transmitted to humans through direct contact with infected birds at various stages like slaughtering, handling, processing, packaging, storing, and decomposing the carcasses, as well as the consumption of contaminated poultry meat [11]. In addition, E. coli often carries multiple drug-resistant plasmids and under stress, they readily transfer those plasmids to other bacteria present in the intestine by horizontal gene transfer mechanisms [12].
E. coli causing diarrhoea in humans carry distinct virulence genes that enable their detection by polymerase chain reaction (PCR), and these genes are used as targets to identify different pathogroups of E. coli in food [13]. These include the astA encoding enterotoxin EAST1 of enteroaggregative E. coli (EAEC) heat-stable enterotoxin (EAST1), bfpA encoding bundle forming pilus of enteropathogenic E. coli (EPEC), ipaHencoding invasion plasmid antigen gene of enteroinvasive E. coli (EIEC), lt and st genes encoding a heat-labile toxin and a heat-stable toxin, respectively of enterotoxigenic E. coli (ETEC), and stx1and stx2 encoding Shiga-toxin1 and 2 of enterohaemorrhagic E. coli (EHEC) [13, 14]. There is a lack of evidence supporting the relationship between E. coli causing gastroenteritis in humans and the multidrug-resistant virulent genes of E. coli isolated from poultry [15]. Hence, identification of the virulence genes by screening diarrheagenic E. coli (DEC) pathotypes is crucial to understand the risk of human infections from these food sources. We hypothesize that since there are very few studies from India about screening the prevalence of diarrheagenic E. coli from chicken faeces [16,17] and without reliable evidence to estimate the antibiotic consumption in livestock, the links between antibiotic consumption and resistance patterns are poorly investigated. To test this hypothesis, the present study is carried out to understand the use of antibiotics in the poultry industry in and around Deralakatte, Mangalore, determine the occurrence of DEC from chicken faces, and study the antibiotic resistance pattern of the obtained isolates.
MATERIALS AND METHODS
Study setting
A prospective observational study was conducted on chicken faeces collected from 30 different chicken farms (broilers, egg-layers and mixed) situated in the Deralakatte suburb of Mangalore, India. A total of 50 chicken faeces samples were collected between April and September 2019.
Data collection from the survey of poultry farms
The knowledge, attitude, and perspectives of farmworkers and managers were assessed towards certain aspects such as management of poultry farms in terms of cleanliness and sanitation, awareness about zoonotic infections, use of antibiotics in poultry and drug resistance. The assessment was done through a structured questionnaire-based survey, randomly collected every week from different farms. The farmworkers or poultry handlers were requested to take up the survey and answer the questions as per their knowledge. Participation was entirely voluntary, and participants were informed about the confidentiality of the information collected. Written informed consent was obtained from each participant. The questionnaire was designed in the English language. It was translated to the farmworkers in the preferred local language (Hindi or Kannada) if they were found having difficulty in understanding or writing in English. The study design was approved by the central ethics committee NITTE (Deemed to be University), Ref: NU/CEC/2018/0175, dated 19/01/2018.
Sample collection, processing and identification of Escherichia coli
Approximately 15 g of fresh faecal droppings were collected each week randomly in a sterile plastic container. Ten grams of the sample was mixed with 90 ml of sterile normal saline (0.85% NaCl) and homogenized. Another 5 g sample was inoculated into selective pre-enrichment trypticase soy broth (TSB) (HiMedia, Mumbai, India). Ten microliters of the homogenized specimen and the pre-enriched sample were cultured onto Eosin methylene blue agar (EMB) and MacConkeyagar (MAC) plates (HiMedia, Mumbai, India). All the plates were incubated at 37ºC for 24 h.
The isolates were identified by standard microbiological techniques such as colony morphology, Gram staining, oxidase reaction from the colony, and standard biochemical reactions (Indole, methyl red, Voges-Proskauer, citrate utilization, urease, triple sugar iron agar test and mannitol motility test) for E. coli as described in the Bergey’s manual [18]. All the confirmed isolates were subcultured onto trypticasesoy agar (TSA) (HiMedia, Mumbai, India) and incubated at 370C for 24 h to yield pure growth. The pure isolated colonies were preserved in Luria Bertani (LB) broth (HiMedia, Mumbai, India) with 30 % glycerol and stored at -20°C for further molecular work.
Antibiotic susceptibility testing
Each confirmed isolate was tested using the standard Kirby-Bauer disk diffusion technique [19] on Muller-Hinton agar (MHA) (HiMedia, Mumbai, India) as recommended by the Clinical and Laboratory Standards Institute (CLSI) 2017 guidelines [20]. A panel of antibiotics (HiMedia, Mumbai, India) used as per CLSI guidelines for E. coli susceptibility testing which included ampicillin (30 ), amikacin (30 µg), amoxyclav (20/10 µg), ceftazidime (30 µg), ceftazidime clavulanic acid (30/10 µg), ceftriaxone (30 µg), cefotaxime (30 µg), cefuroxime (30 µg), cefepime (30 µg), cefoperazone sulbactam (75/15 µg), cotrimoxazole (25 µg), chloramphenicol (30 µg), ciprofloxacin (5 µg), imipenem (10 µg), meropenem (10 µg), ertapenem (10 µg), erythromycin (15 µg), gentamicin (10 µg), tigecycline (15 µg), tetracycline (30 µg) and piperacillin-tazobactam (100/10 µg). The zones of inhibition were measured and interpreted as sensitive or resistant. E. coli ATCC 25922 (USA) was used as a positive control. The multiple antibiotic resistance (MAR) index was determined as the ratio of the total antibiotics used to the number of antibiotics to which the bacterium was resistant [21].
Detection of extended-spectrum beta-lactamase (ESBL) and metallo-b– lactamase (MBL)
Screening of ESBL isolates was done by phenotypic confirmatory disk diffusion test (PCDDT) as described by the CLSI guidelines and Indernath S et al. 2018 [22]. Initially, based on antibiogram analysis, the isolates exhibiting resistance to at least one of the cephalosporin antibiotics such as ceftazidime (30 µg), cefotaxime (30 µg) or ceftriaxone (30 µg), were presumed to be positive for ESBL production. Later to confirm, strains were tested using ceftazidime (30 µg) alone and in combination with inhibitor clavulanic acid (30/10 µg). An isolate was phenotypically confirmed as an ESBL producer when the difference in the zone of inhibition of the drug and the inhibitor was ³ 5 mm compared to the cephalosporin alone [CLSI 2017].
The strains were also confirmed for ESBL by Triple ESBL detection strip Ezy MICTM(MIX+/MIX) (HiMedia, Mumbai, India) as per the manufacturer’s instructions. The upper half of the ESBL detection strip designated as ‘MIX+’ is coated with ceftazidime, cefotaxime and cefepime plus the inhibitor clavulanic acid, while the lower half of the strip designated as ‘MIX’ is coated with the same antibiotics in a reverse concentration gradient, but without an inhibitor. The strip was placed on the MHA plate inoculated by the test bacterium and minimum inhibitory concentrations (MICs) were read from the MIX and MIX+ ends of the strip. The isolate was considered ESBL-positive if the MIX/MIX+ value was ≥ 8 µg/ml.
Detection of metallo-β-lactamase (MBL) production was done using the E-strips (HiMedia, Mumbai, India) containing only meropenem (4-256 µg/ml) at one side and a combination of meropenem with EDTA (1-64 µg/ml) on the other side. MHA plate was inoculated with the test bacterium by lawn and the E-strip was placed in the centre of the inoculated plate. After overnight incubation at 37°C, MBL production was defined when the zone of inhibition ratio of meropenem alone and meropenem + EDTA was ≥ 8 mm. A representative image of ESBL and MBL detection is shown in Figure 1.
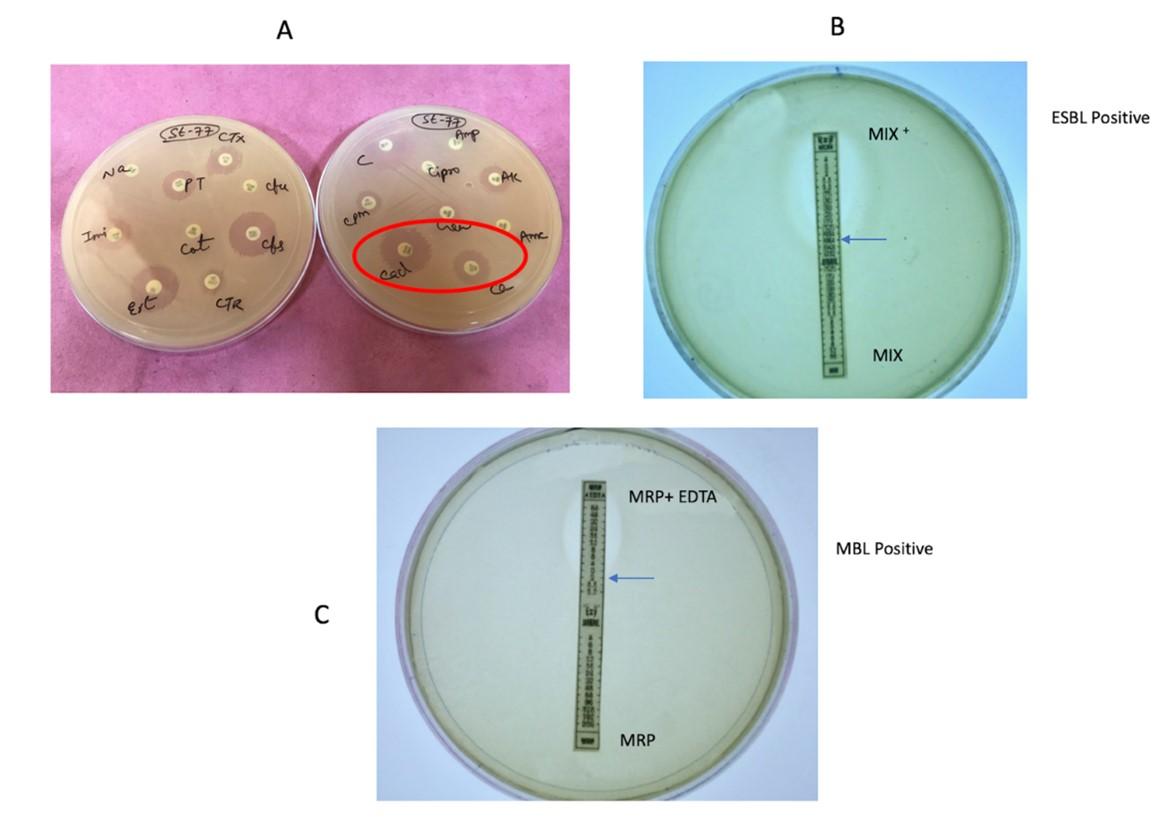
Table1. List of primers used in the study.
DNA extraction and PCR
DNA was extracted using the crude method. E. coli isolates preserved in LB broth with glycerol were subcultured onto fresh MAC. After overnight incubation at 37°C, pure colonies from each plate were inoculated into 5 ml of LB broth and incubated at 37°C for 24 h. The overnight broth was centrifuged at 10,000 rpm for 10 min. The supernatant was discarded, and 50 μl pellet of this culture was added to 450 μl of 1X TE (Tris-HCl and EDTA) buffer. After vortexing, it was incubated at 95°C – 97°C in a dry bath for 15 min and kept on ice for 10 min. The DNA thus obtained was stored at -20°C [23]. Purity and the DNA concentration measured using Nano Drop® spectrophotometer (ND-1000, Thermo Fisher Scientific). DNA concentration between 150-200 ng was used for PCR.
The isolates were tested for the virulence genes astA for the presence of enteroaggregative E. coli (EAEC), bfpA and eaeA gene for enteropathogenic E. coli (EPEC), ipaH gene for enteroinvasive E. coli (EIEC), lt and st gene for enterotoxigenic E. coli (ETEC) and stx 1&stx 2 for enterohaemorrhagic E. coli (EHEC). Previously confirmed isolates with gene sequencing, were used as a positive control for DEC. Distilled water was used everywhere as the negative control. The isolates were also screened for antibiotic resistance genes blaCTX, blaSHV, blaTEM and blaNDM by PCR. For ESBL genes, Klebsiella pneumoniae ATCC 76003 was used as a positive control and for blaNDM, Klebsiella pneumoniae ATCC 2146 was used as the positive control. The primer sequences used in this study and their references are shown in Table-1 [24- 29].
PCR was carried out in a 30 μl reaction mixture containing 10X buffer, 200 mM of dNTPs, 10 pM each of forward and reverse primers, and 1.0 unit of Taq DNA polymerase enzyme. Taq DNA polymerase buffer, dNTPs, and Taq Polymerase and primers were obtained from GeNei, Bangalore, India through Juniper life sciences Pvt. Ltd. Amplification was carried out in an Eppendorf Master cycler nexus GX2 (Thermo Fisher Scientific, Waltham, MA, USA) with an optimized PCR program [27]. Resulting PCR products (10 μl) were mixed with 5 μl of loading dye and analyzed by electrophoresis in 1.5% agarose gel (HiMedia, Mumbai, India) containing 1x Tris-acetate EDTA (1X TAE buffer) and 5 μl of ethidium bromide and visualized in a gel documentation system (Bio-Rad, USA).
Statistical analysis
Antibiotic resistance data were treated as a binary variable (1= susceptible; 2= resistant). Descriptive statistics were performed. The data were analyzed using SPSS (Version 20.0, IBM Corp., Armonk, NY, USA) software and the collected information was summarized using frequency and percentage for qualitative data.
RESULTS
Analysis of data collected from poultry farms
Table 2 highlights the information obtained through the questionnaire-based survey undertaken from 30 different poultry farms. These were broiler chicken farms mainly for meat or mixed layer farms that kept other livestock, including ducks, birds, sheep, pigs, etc., but two were sole egg layer farms. The results highlighted that 18 (60%) farm workers belonged to the age group of 20-40 years and majority of them had education until primary and secondary school. When asked, the workers agreed that they wash their hands frequently and maintain a hygienic environment by cleaning the farms daily. Many participants had adequate knowledge about use of antibiotics in poultry industry and agreed that antibiotics are widely used for prophylaxis and as growth promoters and could also notify the most commonly used antibiotics in their farm were tetracyclines, fluoroquinolones, and colistin but others were unaware of the generic antibiotic names. Complete detail about the survey has been provided in Table 2. Information about antibiotics used has been given in Table 3.
Table 2. Characteristics of 30 chicken farmworkers based on structured questionnaire survey on antibiotic usage in the poultry industry.
Table 3. Details of antibiotics used in the chicken farms obtained through structured questionnaire Survey.
Prevalence of Escherichia coli and antibiotic-resistant pattern
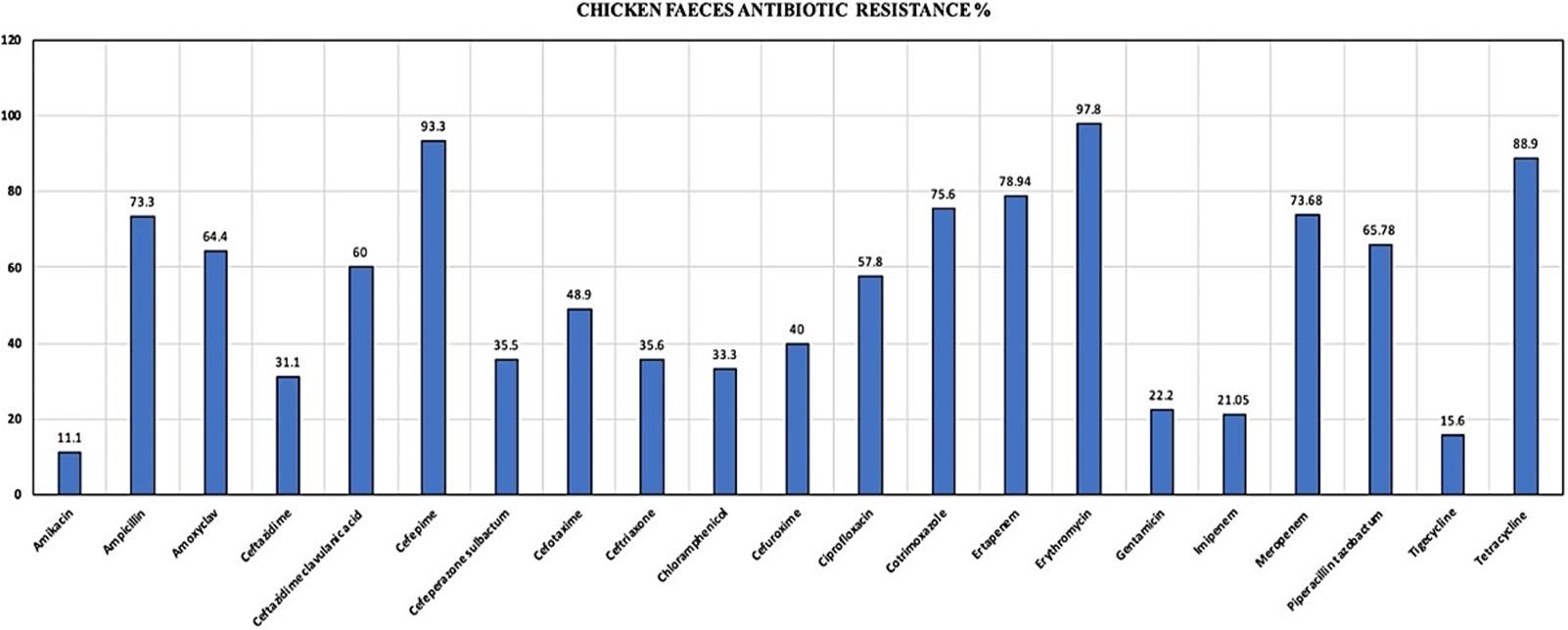
Of chicken faeces samples, 45 (90%) confirmed E. coli strains were isolated. Only one E. coli isolate was selected from one sample for further investigations to prevent duplication of results. There were 7 (15.21%) E. coli isolates, which were positive for phenotypic detection of ESBL. Isolates were highly resistant to cefepime (93.3%), tetracycline (88.9%), ertapenem (78.9%), cotrimoxazole (75.6%), meropenem (73.6%), piperacillin-tazobactam (65.7%), and ciprofloxacin (57.8%) (Figure 2). The resistance pattern proves that the isolates were resistant to fourth generation cephalosporins, carbapenems, and beta-lactam drugs. Resistance to fluoroquinolones was observed in 57.8% of isolates and, 65% of the isolates were also resistant to piperacillin-tazobactam, a beta-lactam inhibitor. There were no isolates that were susceptible to one drug from any three classes of antibiotics. There was no correlation observed between the antibiotic resistance pattern and the type of farm from where the samples were collected.
Prevalence of ESBL and MBL genes
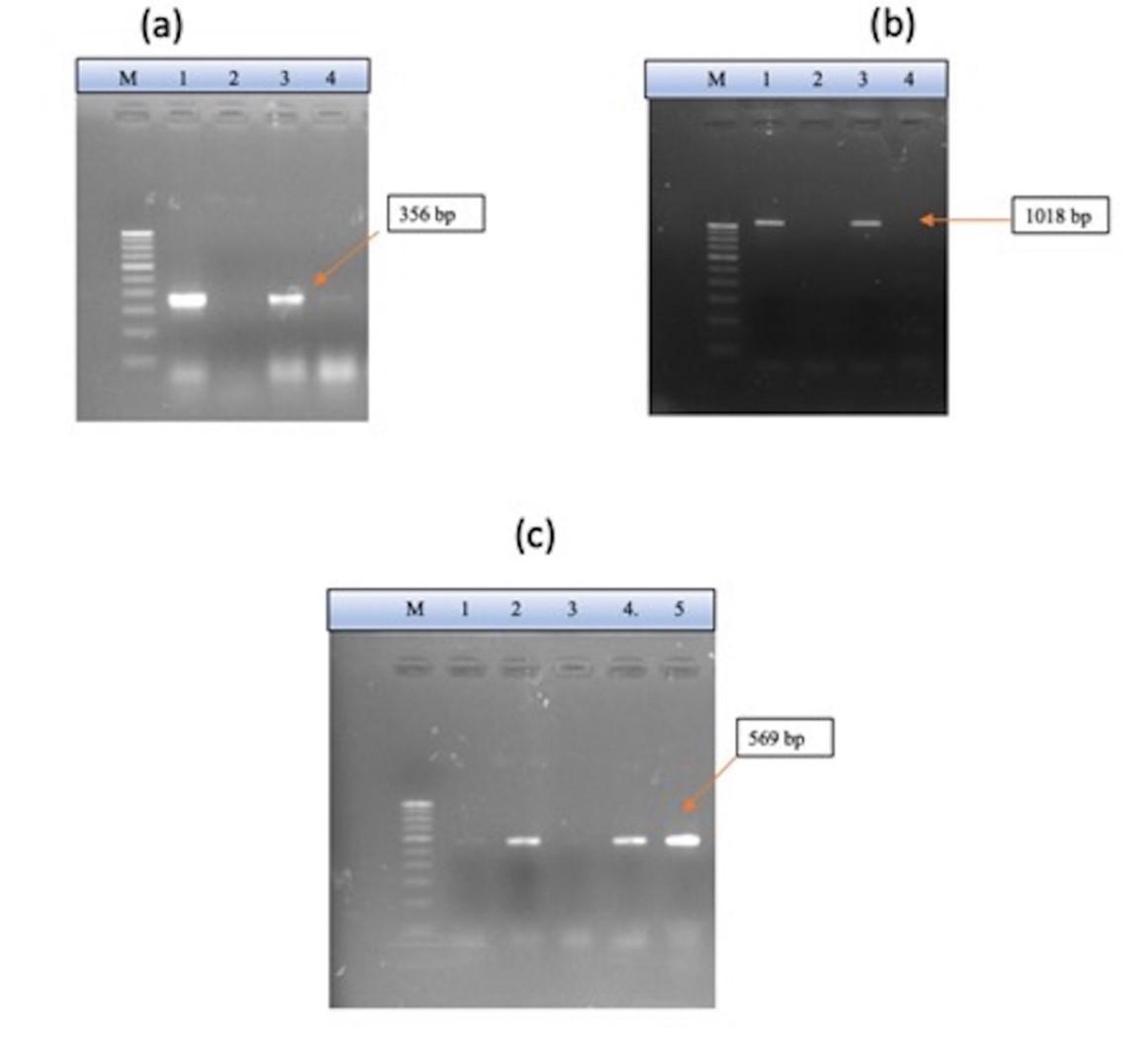
All the isolates were tested for blaCTX, blaSHV, and blaTEM (Figure 3). Although seven isolates were positive for phenotypic detection of ESBL, only a few showed genotypic confirmations by PCR. One isolate was positive for blaCTX, one for blaTEM, and one for the blaSHV gene. The isolate, which was phenotypically detected negative, was confirmed to be positive when tested by PCR. There were no isolates that were positive for all three ESBL genes tested. There was no correlation observed between ESBL detection and types of farms.
Metallo-beta lactamase-encoding gene blaNDM was detected in 12 (26.67%) E. coli isolates. Out of which, eight isolates were susceptible to imipenem, and the rest were resistant. All the 12 isolates exhibited resistance to meropenem and ertapenem.
Prevalence of diarrheagenic Escherichia coli
Figure 4 shows the representative images of DEC pathotypes. Analysis of virulence gene composition revealed presence of astA gene of EAEC in 27 (60%) E. coli isolates out of 45. bfpA and eaeA gene of EPEC were positive for 26 (57.8 %) and 27 (60%) of E. coli isolates respectively. lt gene of ETEC was positive for 12 (12.67%) of the isolates, and the st gene of ETEC was detected in only one isolate (2.22%) of E. coli. ipaH gene of EIEC, stx-1 & stx-2 genes of EHEC was not found in any of the isolates tested. There were 16 (35.6%) isolates, which were positive for both astA and bfpA genes. Hence, the most prevalent type of DEC from this study were the EAEC and EPEC pathotypes. Among EPEC, 17 (37.78%) isolates were positive for both eaeA and bfpA genes together. At the same time, 10(22.22%) isolates were positive only for eaeA and not for the bfpA gene, which means our study reports a high prevalence of typical EPEC over atypical EPEC.
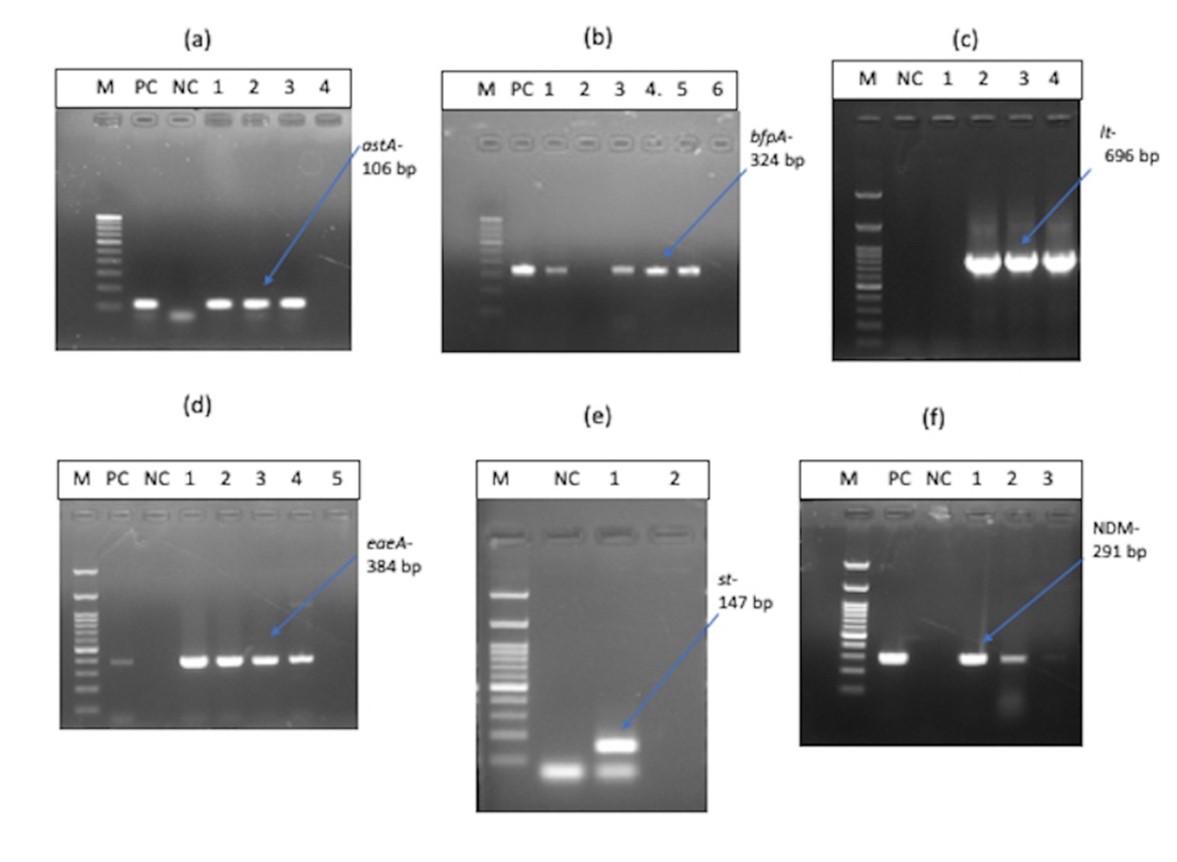
Table 4. Multiple antibiotic resistance (MAR) indices of Escherichia coli isolates from chicken faeces.
Multidrug resistance in Escherichia coli isolates
Of the 45 isolates, 38 (84.44%) were resistant to more than four antibiotics tested with a MAR index of > 0.24, and there were no isolates that were resistant to a minimum of four antibiotics tested (Table 4). Out of the 21 antibiotics tested, two isolates were resistant to 17 (77.27%) antibiotics, and two isolates were resistant to 15 (68.18%) antibiotics. Eight isolates were resistant to 10 (45.45%) antibiotics tested. No isolate was resistant to all the antibiotics, and none of the isolates were susceptible to all the antibiotics tested.
DISCUSSION
This study is the first of its kind from this part of south India, which was undertaken to check the prevalence of multidrug-resistant DEC from chicken faeces using PCR. There are few studies from north India, but their main focus was on Shiga- toxigenic E. coli (STEC) [16, 17, 30]. A study from South Africa shows that DEC’s pathotypes can cause diarrhoea in domestic farm animals, such as chickens depicting its potential for wide circulation and distribution amongst animals and humans [31]. Our study shows a high prevalence (90%) of E. coli isolated from chicken faeces samples. A similar study from Bangladesh has reported a 100% prevalence rate from poultry faeces [32]. However, more studies from India have reported a high prevalence of multidrug-resistant E. coli from poultry, being 51% [33] and 97.9% [34].
There was a low prevalence of ESBL producers from our study, accounting for 15.21%. Contrary to a previous study from north India, which has reported a high prevalence of ESBL-producing E. coli from broiler chickens [35]. Favoring our results, another study from Odisha, India, reported a low prevalence (5.06%) of ESBL-producing E. coli from poultry [36]. It is quite surprising that few isolates that were negative for phenotypic detection showed the presence of ESBL genes when analyzed by molecular technique. This finding shows the need for routine genotypic confirmations for beta-lactamase genes harboring isolates that can easily circulate in the environment and result in multidrug-resistance.
The study isolates were tested for 21 antibiotics and the results showed a high prevalence i.e. 84.44% of multi-drug resistance (MDR) E. coli isolated from chicken faeces. There were 31 (68.8 %) isolates, which were resistant to 10 to 15 antibiotics, 9 (20 %) isolates resistant to 5 to 10 antibiotics, and 5 (11.11 %) isolates that were resistant to 15 to 20 antibiotics. The least resistance was seen in only five isolates. According to the CDC definition, an MDR strain was resistant to at least one drug from three different classes of antibiotics and extreme drug-resistance (XDR) as the one that was resistant to one or more antibiotics from all major classes of antibiotics [37]. This study’s resulting drug resistance pattern alarms the presence of highly drug-resistant enteric pathogens prevalent in farm chickens. When they enter the food chain, humans will become passive carriers of these pathotypes.
In the present study, resistance to carbapenems, the drug of choice for treating multi-drug-resistant beta-lactam positive strains, leaves the health authorities with no alternating treatment option. The high resistance towards carbapenems (ertapenem and meropenem) shows possibility of cross contamination by humans and vice-versa. A study by Kock et al. proves the possible contamination of carbapenem-resistant-Enterobacteriaceae (CRE) between livestock, its environment and humans where, blaNDM gene was located on different plasmids analyzed by multi-locus sequencing technology (MLST) types ST10 and ST156 [38].
In 2006, the European Union restricted antibiotics use in poultry due to evidence of emerging multi-drug resistance, ultimately disseminating in humans [39]. In 2005, fluoroquinolones were prohibited in the USA as resistance was seen in humans and animals, and fluoroquinolones were used as an effective drug for respiratory diseases [40]. The present study also reveals high resistance to fluoroquinolones, which is a concerning issue. According to a study from the University of Minnesota, the emergence of resistance to fluoroquinolones was observed, a drug of choice for treating E. coli infections [41]. Also, it is essential in conditions where resistance to trimethoprim-sulfamethoxazole and extended-spectrum beta-lactams was observed.
The prevalence of DEC varies globally among different locations, regions, and countries. EPEC is the most commonly isolated pathotype of diarrheagenic E. coli. The virulent gene is involved in the expression of bundle forming pilus (bfpA) toxin, which causes acute and chronic watery diarrhoea, especially the atypical EPEC, which causes diarrhoea in domestic animals. EAEC is also associated with acute and persistent diarrhoea. The ipaH gene of EIEC is similar to Shigella spp., responsible for inducing dysentery in humans. STEC infections are rare in India, but few studies proved high chances of their emergence in the future [28]. stx1 and stx2 genes of EHEC or STEC mainly cause hemolytic uremic syndrome. Our study shows that poultry is the leading carrier of the astA gene of EAEC (60%), followed by the bfpA (57.8%) and eaeA (60%) genes of EPEC. A similar result for EPEC was noted in Burkina Faso’s study, accounting for a 37 % prevalence of EPEC and only a 6% prevalence rate for EAEC from chicken faeces samples [14].
The present study mainly highlights the occurrence of EPEC and EAEC pathotypes of DEC from the Deralakatte suburb of Mangalore city, which demands a big-scale investigation on pathotypes of E. coli to be detected in other areas. The outcome of the questionnaire-based survey highlights the regular use of antibiotics in poultry for therapeutic and prophylactic purposes. Though the workers were not aware of the antibiotic’s generic or common names when asked but produced the vials of antibiotics used for poultry. Among frequently used antibiotics, quinolones, penicillin, and tetracyclines were the most commonly used as per the survey (Table 3). A study from Sudan shows results similar to the present study, where the most frequently used antibiotics for animal farming were quinolones and tetracyclines [42]. In the present study, in addition to quinolones and tetracyclines, high resistance was observed for carbapenems and fourth-generation cephalosporin, i.e., cefepime. However, there is no evidence if these antibiotics were used for disease prevention or as a growth promoter.
Limitations of the present study include a brief study period with a smaller number of samples. The survey was conducted only on 30 chicken farms that demand future studies to explore more farms. Despite these limitations, this study delivers adequate knowledge about the high prevalence of EAEC and EPEC from chicken faeces in this rural part of Mangalore.
CONCLUSION
Our data suggested that diarrheagenic E. coli, mainly EAEC and EPEC from the study farms may act as reservoirs of antibiotic drug resistance which may be mobilized into human populations. Survey data suggested that maintaining personal and farm hygiene, cleaning cages/ egg-laying trays/ slaughter areas and providing clean feed will reduce infections among poultry and farmworkers. Finally, there should be public awareness towards restricting the excessive and non-therapeutic use of medically important antibiotics in the poultry industry. There should be programs and policies created by different organizations to work towards achieving better health outcomes which will help achieve a one health approach.
ACKNOWLEDGEMENTS
This research received no external funding. The authors acknowledge gratitude to KS Hegde Medical Academy, NITTE (Deemed to be University), Mangalore for supporting the successful conduct of the research and providing funding support (grant no. NUFR3/2017/06/03) in the form of a Ph.D. Scholarship. The authors thank Mr. Nandith P.B for helping during the survey and communicating with the farmworkers in their understandable local language.
AUTHOR CONTRIBUTIONS
Conceptualization, A.V.S. and S.G.; data curation, S.G., A.V.S., S.H.K. and G.S.K.; formal analysis, S.H.K., A.V.S. and S.G.; investigation, S.G.; methodology, S.G.; project administration, S.G. and A.V.S.; resources, S.G., A.V.S. and G.S.K.; software, S.H.K., A.V.S., S.G. and G.S.K.; supervision, S.H.K. and A.V.S.; writing—original draft, S.G.; writing—review & editing, S.G., S.H.K. and A.V.S. All authors have read and agreed to the published version of the manuscript.
CONFLICTS OF INTEREST
There is no conflict of interest among the authors.
References
- [1][1] Perreten V. Resistance in the food chain and in bacteria from animals: relevance to human infections. Frontiers in antimicrobial resistance: a tribute to Stuart B. Levy. 2005:446-464.
- [2]Jadhav S, Hussain A, Devi S, Kumar A, Parveen S, Gandham N, et al. Virulence characteristics and genetic affinities of multiple drug resistant uropathogenic Escherichia coli from a semi urban locality in India. PloS one. 2011;6(3): e18063.
- [3]Van den Bogaard AE, London N, Driessen CA, Stobberingh EE. Antibiotic resistance of faecal Escherichia coli in poultry, poultry farmers and poultry slaughterers. Journal of Antimicrobial chemotherapy. 2001;47(6):763-771.
- [4]Xiong W, Wang Y, Sun Y, Ma L, Zeng Q, Jiang X, et al. Antibiotic-mediated changes in the fecal microbiome of broiler chickens define the incidence of antibiotic resistance genes. Microbiome. 2018;6(1):1-1.
- [5]Landers TF, Cohen B, Wittum TE, Larson EL. A review of antibiotic use in food animals: perspective, policy, and potential. Public health reports. 2012;127(1):4-22.
- [6]Van Boeckel TP, Brower C, Gilbert M, Grenfell BT, Levin SA, Robinson TP, et al. Global trends in antimicrobial use in food animals. Proceedings of the National Academy of Sciences. 2015;112(18):5649-5654.
- [7]Mubito E, Shahada F, Kimanya M, Buza J. Antimicrobial use in the poultry industry in Dar-es-Salaam, Tanzania and public health implications. 2014.
- [8]van den Bogaard AE, Stobberingh EE. Antibiotic usage in animals. Drugs. 1999;58(4):589-607.
- [9]Rugumisa BT, Call DR, Mwanyika GO, Mrutu RI, Luanda CM, Lyimo BM, et al. Prevalence of antibiotic-resistant fecal Escherichia coli isolates from penned broiler and scavenging local chickens in arusha, Tanzania. Journal of food protection. 2016;79(8):1424-1429.
- [10]Ibrahim RA, Cryer TL, Lafi SQ, Basha EA, Good L, Tarazi YH. Identification of Escherichia coli from broiler chickens in Jordan, their antimicrobial resistance, gene characterization and the associated risk factors. BMC veterinary research. 2019;15(1):1-6.
- [11]Kuldeep D, Sandip C, Barathidasan R, Ruchi T, Rajagunalan S, Singh SD. Escherichia coli, an economically important avian pathogen, its disease manifestations, diagnosis and control, and public health significance: a review. Research Opinions in Animal and Veterinary Sciences. 2013;3(6):179-194.
- [12]Amer MM, Mekky HM, Amer AM, Fedawy HS. Antimicrobial resistance genes in pathogenic Escherichia coli isolated from diseased broiler chickens in Egypt and their relationship with the phenotypic resistance characteristics. Veterinary world. 2018;11(8):1082.
- [13]aton AW, Paton JC. Detection and characterization of Shiga toxigenic Escherichia coli by using multiplex PCR assays for stx 1, stx 2, eaeA, enterohemorrhagic E. coli hlyA, rfb O111, and rfb O157. Journal of clinical microbiology. 1998;36(2):598-602.
- [14]Kagambega A, Martikainen O, Siitonen A, Traore AS, Barro N, Haukka K. Prevalence of diarrheagenic Escherichia coli virulence genes in the feces of slaughtered cattle, chickens, and pigs in Burkina Faso. MicrobiologyOpen. 2012;1(3):276-284.
- [15]Zhang J, Massow A, Stanley M, Papariella M, Chen X, Kraft B, et al. Contamination rates and antimicrobial resistance in Enterococcus spp., Escherichia coli, and Salmonella isolated from “No antibiotics added”–labeled chicken products. Foodborne pathogens and disease. 2011;8(11):1147-1152.
- [16]Wani SA, Samanta I, Bhat MA, Nishikawa Y. Investigation of shiga toxin‐producing Escherichia coli in avian species in India. Letters in applied microbiology. 2004;39(5):389-394.
- [17]Dutta TK, Roychoudhury P, Bandyopadhyay S, Wani SA, Hussain I. Detection & characterization of Shiga toxin producing Escherichia coli (STEC) & enteropathogenic Escherichia coli (EPEC) in poultry birds with diarrhoea. The Indian journal of medical research. 2011;133(5):541.
- [18]Forbes, B.A., Sahm, D.F., Weissfeld, A.S., Bailey, W.R.Bailey & Scott’s Diagnostic Microbiology. 12th Edition, Elsevier Mosby, St. Louis. 2007.
- [19]Bauer AW. Antibiotic susceptibility testing by a standardized single disc method. Am J clinpathol.1966;45:149-158.
- [20]CLSI.Performance standards for Antimicrobial Susceptibility Testing; 27th ed. CLSI Supplement M100.Wayne, PA: Clinical and Laboratory Standards Institute; 2017.
- [21]Krumperman PH. Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of fecal contamination of foods. Applied and environmental microbiology. 1983;46(1):165-170.
- [22]Indernath S, Priyadharsini RI, Manikkanan M, Babu SV. Prevalence and phenotypic characterization of ESBL producing E. coli and Klebsiella among the fecal isolates of normal population. Indian J Microbiol Res. 2018;5(1):52-6.
- [23]Nisha KV, Veena SA, Rathika SD, Vijaya SM, Avinash SK. Antimicrobial susceptibility, risk factors and prevalence of bla cefotaximase, temoneira, and sulfhydryl variable genes among Escherichia coli in community-acquired pediatric urinary tract infection. Journal of laboratory physicians. 2017;9(03):156-162.
- [24]Colomer-Lluch M, Jofre J, Muniesa M. Antibiotic resistance genes in the bacteriophage DNA fraction of environmental samples. PloS one. 2011;6(3):e17549.
- [25]Jemima SA, Verghese S. Multiplex PCR for blaCTX-M & blaSHV in the extended spectrum beta lactamase (ESBL) producing gram-negative isolates. Indian Journal of Medical Research. 2008;128(3):313.
- [26]Teo J, Ngan G, Balm M, Jureen R, Krishnan P, Lin R. Molecular characterization of NDM-1 producing Enterobacteriaceae isolates in Singapore hospitals. Western Pacific surveillance and response journal: WPSAR. 2012;3(1):19.
- [27]Bisi-Johnson MA, Obi CL, Vasaikar SD, Baba KA, Hattori T. Molecular basis of virulence in clinical isolates of Escherichia coli and Salmonella species from a tertiary hospital in the Eastern Cape, South Africa. Gut pathogens. 2011;3(1):1-8.
- [28]Shetty VA, Kumar SH, Shetty AK, Karunasagar I, Karunasagar I. Prevalence and characterization of diarrheagenic Escherichia coli isolated from adults and children in Mangalore, India. Journal of laboratory physicians. 2012;4(01):024-029.
- [29]Binet R, Deer DM, Uhlfelder SJ. Rapid detection of Shigella and enteroinvasive Escherichia coli in produce enrichments by a conventional multiplex PCR assay. Food microbiology. 2014; 40:48-54.
- [30]Debbarma S, Bora DP, Hazarika RA, Tamuly S, Barua AG, Das P, et al. Molecular characterization of Escherichia Coli isolates from food animals. Indian Journal of Animal Research. 2020;54(8):985-993.
- [31]Aijuka M, Buys EM. Persistence of foodborne diarrheagenic Escherichia coli in the agricultural and food production environment: Implications for food safety and public health. Food microbiology. 2019; 82:363-370.
- [32]Bashar T, Rahman M, Rabbi FA, Noor R, Rahman MM. Enterotoxin profiling and antibiogram of Escherichia coli isolated from poultry feces in Dhaka district of Bangladesh. Stamford journal of microbiology. 2011;1(1):51-57.
- [33]Hussain A, Shaik S, Ranjan A, Nandanwar N, Tiwari SK, Majid M, et al. Risk of transmission of antimicrobial resistant Escherichia coli from commercial broiler and free-range retail chicken in India. Frontiers in microbiology. 2017; 8:2120.
- [34]Saharan VV, Verma P, Singh AP. Escherichia coli, Salmonella spp., and Staphylococcus aureus susceptibility to antimicrobials of human and veterinary importance in poultry sector of India. Journal of Food Safety. 2020;40(1): e12742.
- [35]Brower CH, Mandal S, Hayer S, Sran M, Zehra A, Patel SJ, et al. The prevalence of extended-spectrum beta-lactamase-producing multidrug-resistant Escherichia coli in poultry chickens and variation according to farming practices in Punjab, India. Environmental health perspectives. 2017;125(7):077015.
- [36]Kar D, Bandyopadhyay S, Bhattacharyya D, Samanta I, Mahanti A, Nanda PK, et al. Molecular and phylogenetic characterization of multidrug resistant extended spectrum beta-lactamase producing Escherichia coli isolated from poultry and cattle in Odisha, India. Infection, Genetics and Evolution. 2015; 29:82-90.
- [37]Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clinical microbiology and infection. 2012;18(3):268-281.
- [38]Köck R, Daniels-Haardt I, Becker K, Mellmann A, Friedrich AW, Mevius D, Schwarz S, Jurke A. Carbapenem-resistant Enterobacteriaceae in wildlife, food-producing, and companion animals: a systematic review. Clinical Microbiology and Infection. 2018;24(12):1241-50.
- [39]EU R. Ban on antibiotics as growth promoters in animal feed enters into effect. Brussels: Europa. 2005.
- [40]Nelson JM, Chiller TM, Powers JH, Angulo FJ. Fluoroquinolone-resistant Campylobacter species and the withdrawal of fluoroquinolones from use in poultry: a public health success story. Clinical Infectious Diseases. 2007;44(7):977-980.
- [41]ohnson JR, Kuskowski MA, Menard M, Gajewski A, Xercavins M, Garau J. Similarity between human and chicken Escherichia coli isolates in relation to ciprofloxacin resistance status. The Journal of infectious diseases. 2006;194(1):71-78.
- [42]Eltayb A, Barakat S, Marrone G, Shaddad S, StålsbyLundborg C. Antibiotic use and resistance in animal farming: a quantitative and qualitative study on knowledge and practices among farmers in Khartoum, Sudan. Zoonoses and Public Health. 2012;59(5):330-338.